Subscribe to Pittwire Today
Get the most interesting and important stories from the University of Pittsburgh.Researcher Creates Enzymes From Scratch
This story, written by Alla Katsnelson, originally appeared in the fall 2018 issue of Pitt Med magazine.
Enzymes are the workhorses of the cell — and of all biological systems. These specialized proteins set into motion all the cells’ chemical reactions, making sure they run fast enough for life on a biochemical level to proceed. Each enzyme precisely aligns with the cellular processes it regulates. Thanks to evolution, our world is full of these meticulously well-suited proteins.
A study published earlier this year by Ann Donnelly, a research specialist in the Department of Biomedical Informatics, has revealed that scientists can also make working enzymes from scratch.
The study was published in January in Nature Chemical Biology. Donnelly, who came to Pitt in 2017, did the work as a PhD student in the lab of Michael Hecht at Princeton.
“We showed that you can take novel protein sequences that nature has never seen before and put them in natural systems — and they can function,” Donnelly explains.
The findings hint at some fascinating dimensions of our primordial history, she says: Namely, the reactions that governed early cellular processes were much more flexible than they are now. “The enzymes we see today have a lot of evolutionary baggage and have been really refined to do the things they do,” Donnelly says.
But evolution’s solutions weren’t the only ones, it turns out. “Our work suggests it’s possible to replace what we have now with something completely different.”
The synthetic enzyme Syn-F4 was one of a big batch made in Hecht’s lab a decade ago. The group routinely produces synthetic proteins, designing them to conform to a folding pattern called a four-helix bundle and then testing them in mutated strains of the bacterium Escherichia coli. The idea is to see whether any of the synthetic enzymes they make can replace the functions of E. coli genes that have been knocked out. And sometimes the artificial versions do work. Mostly they come into play in a pinch — by switching on cellular processes that can have functions similar to what’s called for.
“But the case with Syn-F4 was a little different,” Donnelly says.
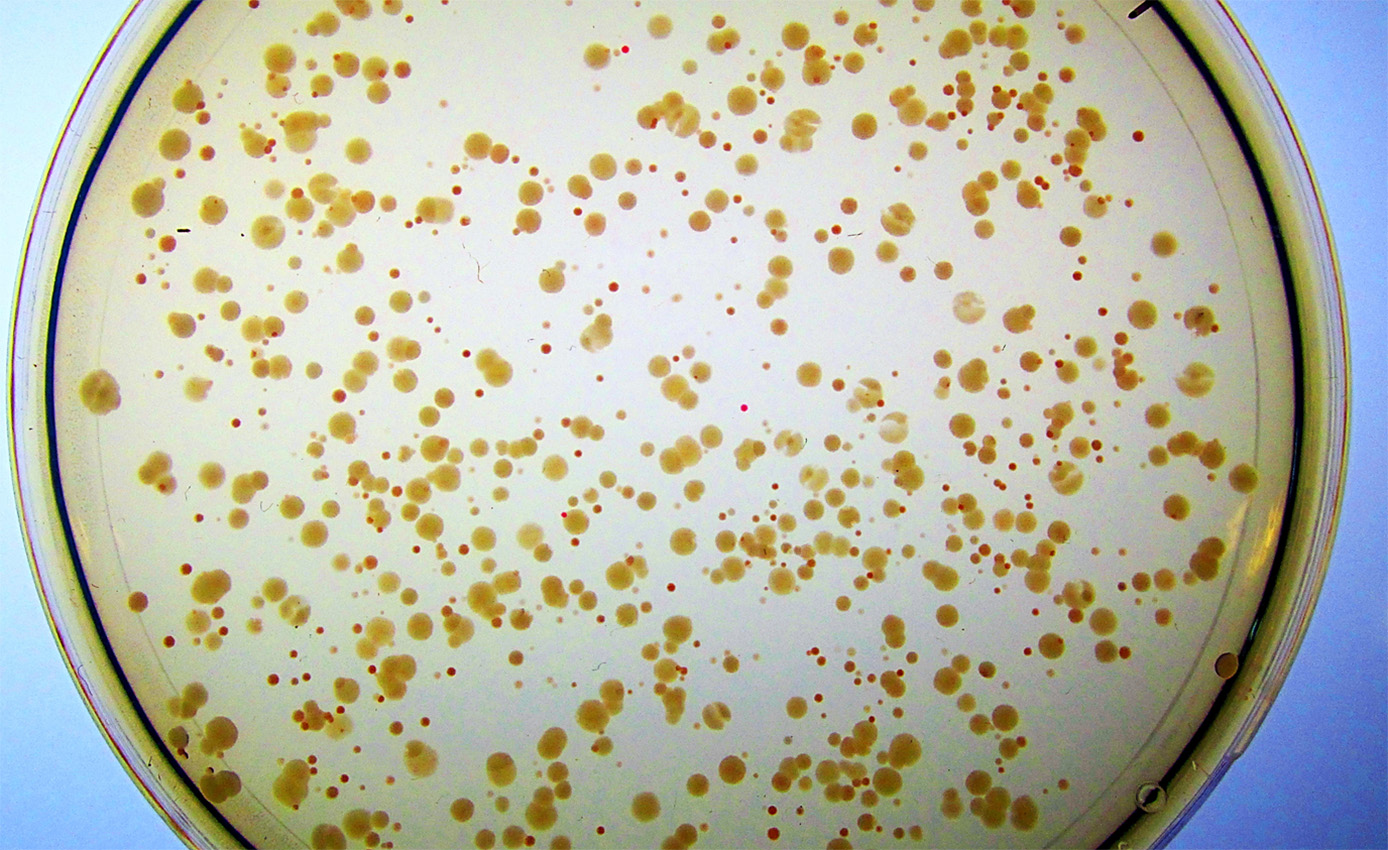
Syn-F4 and its synthetic cohort were evolved to fill in for an E. coli enzyme called Fes that had been hobbled by a mutation. The job of Fes is to release iron from a compound in E. coli that nabs the metal from the environment so that it can be used for healthy growth in the cell. Without Fes, the bacterial colonies grow poorly, speckling with red as iron builds up around them. But when Donnelly added Syn-F4 to these sickly colonies, the red began to disappear, returning the E. coli to its healthy state. “It was clear as day,” she says. “It was unbelievable for me to see this happening in real time.”
So unbelievable, in fact, that she kept mum about it until she had repeated the finding several times. In addition to testing the synthetic protein in living bacteria, she also mixed it directly with its iron-grabbing substrate and biochemically analyzed the ensuing reaction.
Later, she tweaked the substrate by chemically reversing its orientation. She found that this prevented Syn-F4 from working its magic, demonstrating its specificity and supporting the idea that it was working as an enzyme.
It was unbelievable for me to see this happening in real time.
Ann Donnelly, research specialist
What’s interesting, says Donnelly, is that the natural enzyme and the artificial one look completely different. The natural one is about four times as big, and it is known to connect to the substrate through a site that includes the amino acid serine. The artificial enzyme, though, has no serine residue in it whatsoever.
“It’s difficult to pinpoint exactly how it’s working, but at minimum we know that they are not catalyzing the reaction the same way.”
These days at Pitt, in the lab of Erik Wright, an assistant professor of biomedical informatics, Donnelly’s using her ingenuity to focus on a new challenge: understanding how pathogens evolve to become resistant to antibiotics.